|
Bacteria are prokaryotes, i.e. single-celled organisms that lack a nucleus and specialized organelles. They can be broadly classified into two large groups based on their Gram stain (Figure 1), and come in various shapes and sizes (Figure 2). 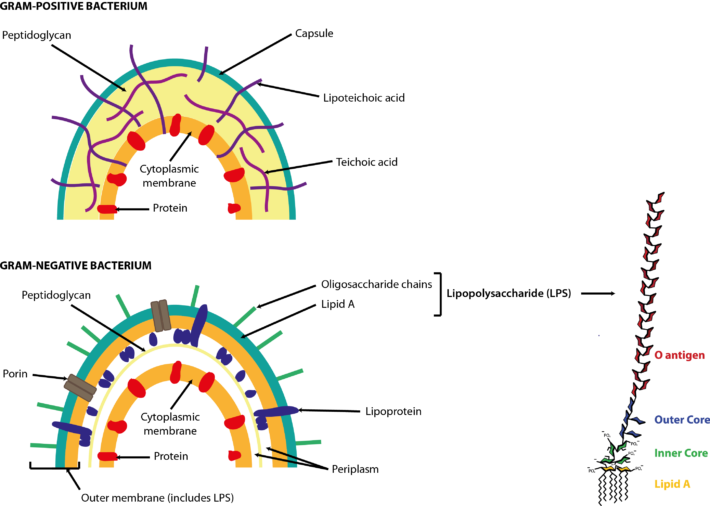 Figure 1. Cell walls of Gram-positive and Gram-negative bacteria. Both types of bacteria can be encapsulated or unencapsulated. Lipopolysaccharide (LPS) on the cell wall of Gram-negative bacteria is considered an endotoxin, and circulating levels of it within the body are associated with metabolic endotoxaemia and inflammation (Cani et al., 2008). The image of the detailed structure of LPS (by Mike Jones) was taken from Wikipedia and is available under a Creative Commons Attribution-Share Alike 3.0 Unported license, via Wikimedia Commons. Over 1000 different species of bacteria have been isolated from the human gastrointestinal tract, with bacteria belonging to the phyla Actinobacteria, Bacteroidetes, Firmicutes and Proteobacteria most predominant and diverse in the faeces of healthy adults (Rajilić-Stojanović et al., 2014; Browne et al., 2016; Lagier et al., 2016). They are by far the most abundant organisms inhabiting the human gastrointestinal tract; consequently, most gut microbiota studies focus on the characterization of bacteria. The gut bacterial microbiota has been referred to recently as the bacteriome, though this should not be confused with the insect bacteriome, a specialized organ that hosts endosymbiotic bacteria. 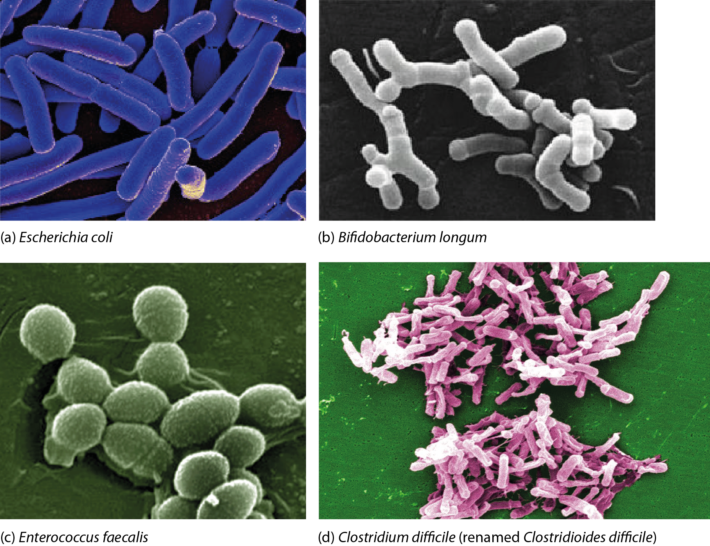 FIgure 2. Electron micrographs of bacteria commonly associated with the human gut microbiota showing some of the shapes of different bacteria. Other examples can be found in the NIAID and Sanofi Pasteur photo albums from which (a) and (d), respectively, were taken. Note that Clostridium difficile has been renamed ‘Clostridioides difficile‘ (Lawson et al., 2016). (b) By Julie6301, available under a Creative Commons Attribution-Share Alike 3.0 Unported license, via Wikimedia Commons. (c) Available under a Creative Commons Attribution-Share Alike 3.0 Unported license, via Wikimedia Commons. Photo credit: Janice Haney Carr Content Providers(s): CDC/ Pete Wardell derivative work: F. Lamiot. References Browne, H. P., Forster, S. C., Anonye, B. O., Kumar, N., Neville, B. A., Stares, M. D., Goulding, D. & Lawley, T. D. (2016). Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature 533, 543-546. Cani, P. D., Bibiloni, R., Knauf, C., Waget, A., Neyrinck, A. M., Delzenne, N. M. & Burcelin, R. (2008). Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 57, 1470-1481. Lagier, J. C., Khelaifia, S., Alou, M. T., Ndongo, S., Dione, N., Hugon, P., Caputo, A., Cadoret, F., Traore, S. I., Seck, E. H., Dubourg, G., Durand, G., Mourembou, G., Guilhot, E., Togo, A., Bellali, S., Bachar, D., Cassir, N., Bittar, F., Delerce, J., Mailhe, M., Ricaboni, D., Bilen, M., Dangui Nieko, N. P., Dia Badiane, N. M., Valles, C., Mouelhi, D., Diop, K., Million, M., Musso, D., Abrahão, J., Azhar, E. I., Bibi, F., Yasir, M., Diallo, A., Sokhna, C., Djossou, F., Vitton, V., Robert, C., Rolain, J. M., La Scola, B., Fournier, P. E., Levasseur, A. & Raoult, D. (2016). Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol 1, 16203. Lawson, P. A., Citron, D. M., Tyrrell, K. L. & Finegold, S. M. (2016). Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O’Toole 1935) Prévot 1938. Anaerobe 40, 95-99. Rajilić-Stojanović, M. & de Vos, W. M. (2014). The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev 38, 996-1047. Comments are closed.
|
Lesley HoylesProfessor of Microbiome and Systems Biology, Nottingham Trent University ArchivesCategories
All
|
